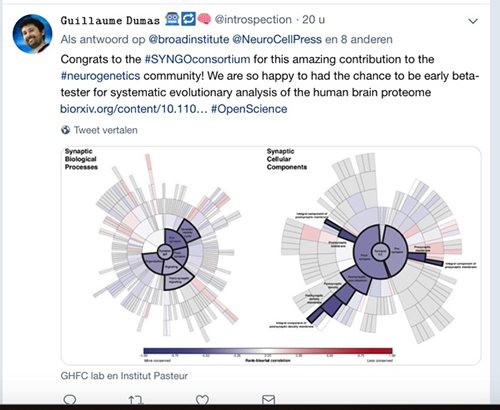
The SYNGO Consortium, coordinated by CNCR scientists Frank Koopmans, Guus Smit and Matthijs Verhage, release the first deep annotation of synaptic genes and publish 1st paper in Neuron
Synapses, the junctions that serve as specialized contacts between nerve cells, are the brain’s fundamental information processing units. A loss of coordinated activity at the synapse lies at the root of many brain disorders (collectively called ‘synaptopathies’). Systematic information resources for the synapse are currently lacking. SYNGO, a collaboration bringing together 15 laboratories worldwide and the Gene Ontology (GO) Consortium, has released SYNGO1.0, the first version of a knowledge base* that aims to represent the neuroscience community’s current scientific knowledge about the genetic architecture of the synapse. Using structured frameworks called ontologies, SYNGO has released nearly 3,000 descriptions of more than 1,100 unique synaptic genes, compiling published experimental information about their protein products’ localization and/or function. This compiled information is both human-readable and machine-readable. SYNGO is fully integrated in the GO knowledgebase, the world’s largest source of information on the functions of genes.
The release of SYNGO 1.0 received press coverage around the world, including the USA, The Netherlands, Singapore, Canada, Italy, Germany and Spain.

In an accompanying paper in the journal Neuron. Consortium members use SYNGO 1.0 to show that synaptic genes have changed little over the course of evolution (that is, are evolutionarily well-conserved), and are functionally much more sensitive (that is, less tolerant) to mutations than other genes expressed in the brain. In addition, the authors also show that variations in many synaptic genes are significantly associated with intelligence, educational attainment, ADHD, autism, and bipolar disorder. Synaptic genes are also much more likely than other brain-expressed genes to bear mutations associated with neuropsychiatric disorders.
The SYNGO Consortium was established in 2015 by Steven Hyman and Guoping Feng of the Stanley Center for Psychiatric Research at the Broad Institute of MIT in Cambridge (MA) and coordinated by Frank Koopmans, Guus Smit and Matthijs Verhage of CNCR, and for the GO Consortium, Paul Thomas at the University of Southern California. Pim van Nierop, former CNCR PI, helped design the project, contributed many annotations and performed quality control. CNCR scientists of the CTG department, Kyoko Watanabe and Danielle Posthuma performed over-representation studies in genome-wide association data on human brain disorders. SYNGO builds on prior initiatives generated in European synapse research consortia (i.e., EU-Synapse, EuroSpin, SynSys).

(1) a standardized framework of definitions (an ontology) for describing the functions, locations, and relationships of proteins and genes in the context of the synapse
(2) expert-curated, literature-based annotations linking synaptic genes and proteins to specific terms
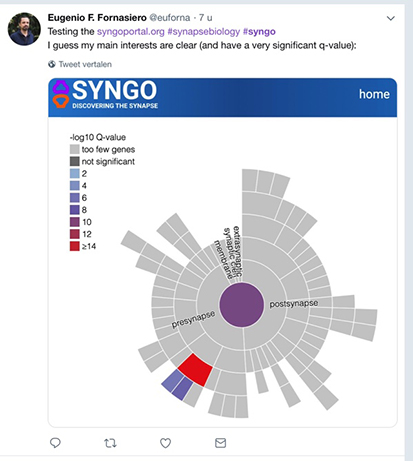
(3) online analysis and visualization tools for evaluating the locations and functions of individual synaptic genes or to perform ‘enrichment’ studies
The SYNGO knowledge base allows researchers to:
(1) search for synaptic proteins, find related proteins, evaluate the evidence for their synaptic localization and/or function and use links to other information on synaptic proteins/genes
(2) analyze gene/protein sets from your -omics experiments and discover how such data is structured, which proteins are synaptic, whether synaptic proteins are over-represented in such data, where they localize in the synapse and what they may do. SYNGO webtools helps users to explore their data, and visualize the analysis results. SYNGO uses a machine-searchable format, fully integrated in the widely-used GO framework
(3) upload new annotations for synaptic genes/proteins using the annotation interface
SYNGO is a unique resource:
(1) SYNGO is collectively supported by the international synapse research community.
(2) SYNGO is the first all-inclusive ontology of the synapse. It represents all synaptic localizations and all aspects of synaptic function in a single ontology in an unbiased manner
(3) SYNGO is built exclusively on evidence from published experiments that can be evaluated by users
(4) intuitive webtools provide advanced analysis and visualization opportunities