
An international group of scientists including researchers of the CNCR have detected 22 independent genetic risk variants for schizophrenia. Thirteen of these have never been reported before. In addition, these novel results point to an enrichment of significant findings in genes that influence L-type calcium channels.
Schizophrenia is a severe mental disorder with a lifetime prevalence of 1%. Current treatments are insufficient. The development of novel treatments is seriously hampered by a lack of etiological insight. Genetic information of more than 20,000 patients and 40,000 healthy controls were investigated. This study is the largest genome-wide study for schizophrenia to date. The scientist used the Swedish National sample for schizophrenia and data-resources from the Psychiatric Genomics Consortium.
Milestone
“This study is an important milestone in understanding the genetic architecture of schizophrenia‚, explains PhD student Christiaan de Leeuw (CNCR). He conducted the gene-set analyses for the study. “For the first time we report association with multiple genes that influence calcium signalling. This is an exciting finding. It provides novel insight into the underlying biology.‚
Schizophrenia is – like many other diseases – a complex disorder. It is influenced by thousands of genetic (and environmental) risk factors. “This study clearly shows that increasing the sample size pays off and will eventually lead to detecting more risk variants‚, says Danielle Posthuma (CNCR). “As the effect of each single risk variant is very small, biological annotation and subsequent gene-set analyses is crucial for interpretation‚, Guus Smit (CNCR) adds.
Synaptic function
It also appears that three sets of genes involved in specific synaptic functions are significantly associated with the risk for schizophrenia. “This result replicates our 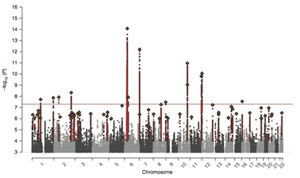
Prof. Patrick Sullivan from the University of North Carolina, Chapel Hill (US) led the study. Sullivan worked at the CNCR in the past year on an honorary professorship from the Royal Academy of Arts and Sciences, The Netherlands.
Genetic Cluster Computer

The study involved an impressive amount of data and required an enormous amount of computing power. This was facilitated by the Dutch Genetic Cluster Computer, which is supported by SURFsara, the VU and the Netherlands Organisation for Scientific Studies (NWO).
The study was published in Nature Genetics, August 25, 2013.
Link: http://www.nature.com/ng/journal/vaop/ncurrent/pdf/ng.2742.pdf