A collaboration between CNCR-FGA and University of Heidelberg shows that protein instability is the generalizable, primary cause. A new prediction tool outperforms all existing predictors. The paper is out now in Biological Psychiatry.
Pathogenic variants in STXBP1/Munc18-1 cause severe encephalopathies that are among the most common in genetic neurodevelopmental disorders. Different molecular disease mechanisms have been proposed and pathogenicity prediction is limited. The new study led by Annemiek van Berkel (FGA), Timon André and Gurdeep Singh (both University of Heidelberg) aimed to define a generalized disease concept for STXBP1-related disorders and improve prediction.
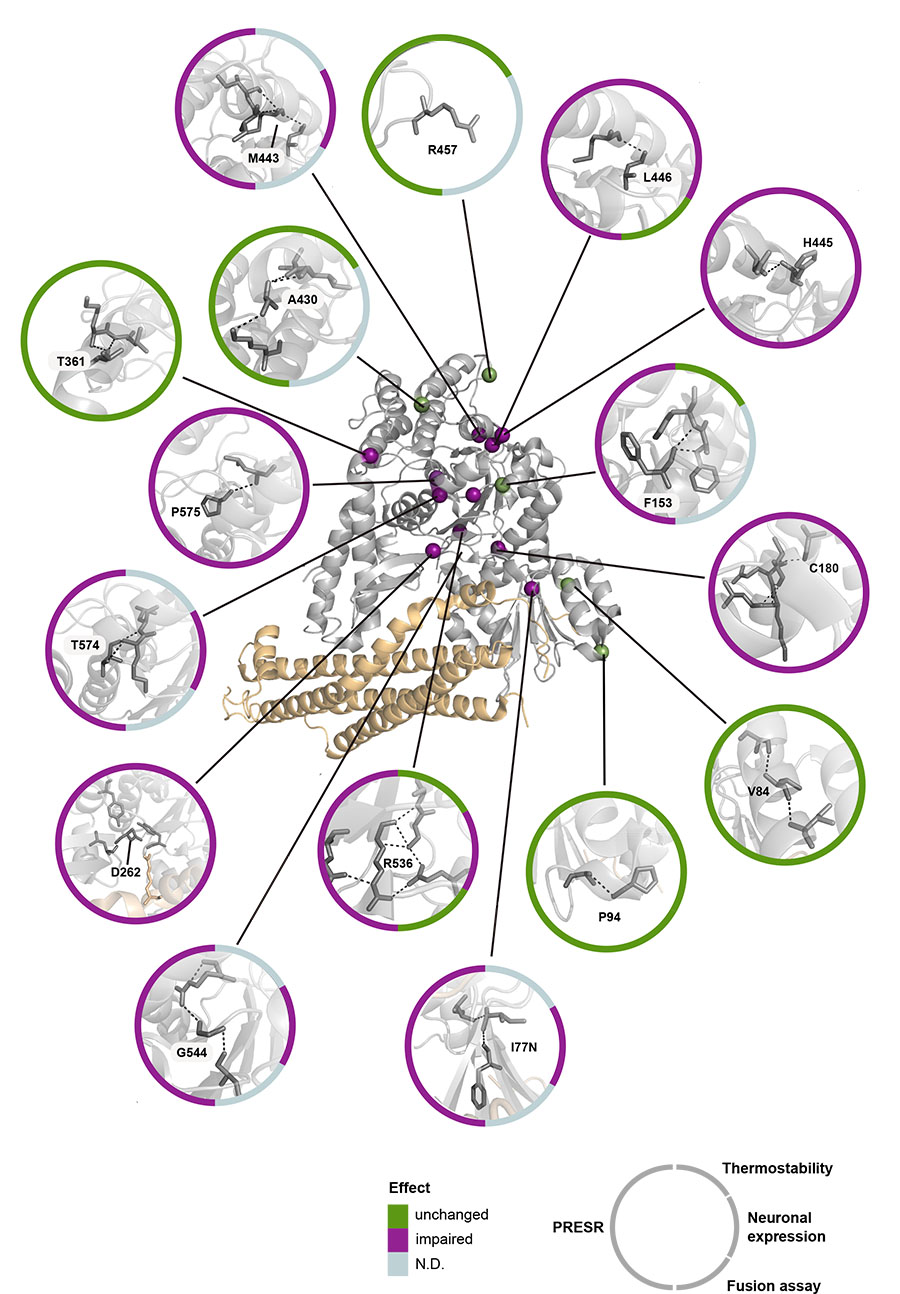
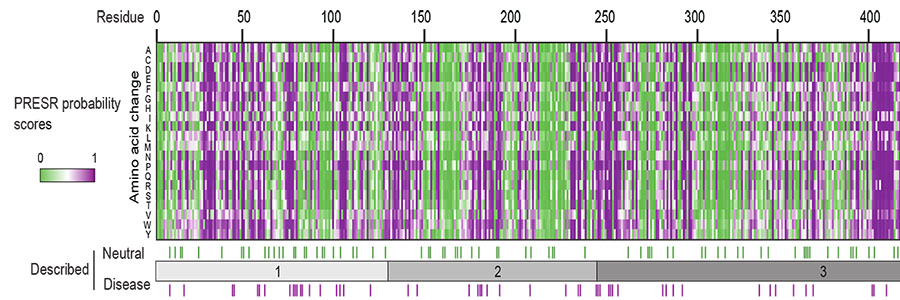
A cohort of 11 disease-associated and five neutral variants (detected in healthy individuals) was tested in three cell-free assays, and in heterologous cells and primary neurons. A machine learning algorithm (PRESR) that uses both sequence- and 3D structure-based features was developed to improve pathogenicity prediction using 231 known disease-associated variants and comparison to our experimental data.
The researchers found that disease-associated, but none of the neutral variants produced reduced protein levels. Cell-free assays demonstrated directly that disease-associated variants have reduced thermostability, with most variants denaturing around body temperature. In addition, most disease-associated variants impaired SNARE- mediated membrane fusion in a reconstituted assay. Moreover, PRESR outperformed existing tools substantially: Matthews correlation coefficient = 0.71 versus <0.55.
Taken together, this study establishes intrinsic protein instability as the generalizable, primary cause for STXBP1-related disorders and shows that protein-specific ortholog and 3D information improves disease prediction. PRESR is a publicly available diagnostic tool:
The paper is available here.
The prediction tool is available here: PRESR.russelllab.org