Rick Jansen and Klaus Linkenkaer-Hansen (INF) and collaborators published their finding of candidate genes underlying amplitude, frequency or interactions of hippocampal oscillations in PLoS ONE.
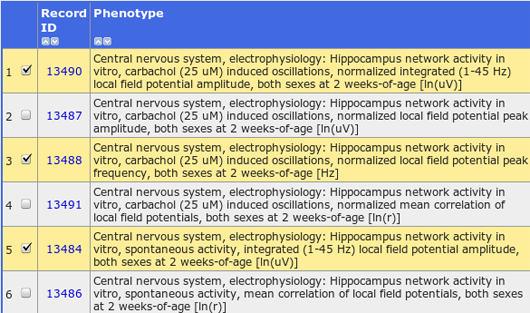
Anxious behavior, spatial orientation and memory have been linked to gamma oscillations in the hippocampus. In this study, Jansen and Linkenkaer-Hansen performed the first genome-wide search for genes influencing hippocampal oscillations. Oscillations were pharmacologically induced in hippocampal slices from a population of 29 BXD recombinant inbred mouse strains. The oscillations were characterized by their amplitude, frequency and interactions, and related to genotypic and gene-expression variation of the mouse strains.
Eight candidate genes were identified, including Plcb1, a phospholipase that is known to influence hippocampal oscillations. Two genes coding for calcium channels, Cacna1b and Cacna1e, which mediate presynaptic transmitter release and were not known to regulate hippocampal network activity, were also identified.
It was further addressed whether genetic predisposition for having strong, fast or tightly coupled hippocampal oscillations affects behavior. Genetic correlations between the hippocampal traits and traits from the GeneNetwork database suggest that mice generating large-amplitude oscillations move less and linger more in a novel environment. Hippocampal oscillations may mediate the effect of the identified genes onto these behavioral traits.
Novel Candidate Genes Associated with Hippocampal Oscillations
Rick Jansen [1,2], Jaap Timmerman [2], Maarten Loos [3], Sabine Spijker [3], Arjen van Ooyen [2], Arjen B. Brussaard [2], Huibert D. Mansvelder [2], The Neuro-Bsik Mouse Phenomics consortium, August B. Smit [3], Mathisca de Gunst [1], Klaus Linkenkaer-Hansen [2]
1 Department of Mathematics (VU)
2 Department of Integrative Neurophysiology (CNCR)
3 Department of Molecular and Cellular Neurobiology (CNCR)
Traits from this study can now be accessed at www.genenetwork.org, a large database of traits measured on BXD strains: