PhD student Mats Nagel from CTGlab publishes in Nature Communications
Through genome-wide association studies (GWAS), geneticists aim to identify genetic variants that are associated with human traits, such as cognitive function, depression, or personality. Generally, identified variants have very small effects, supporting the idea that many human traits are genetically highly complex. To date, researchers tend to use composite scores to measure traits in the psychological domain. These composite scores summarize information contained in multiple items or symptoms, and have proven very useful in daily practice, e.g., in directing therapeutic intervention or predicting school/job performance. However, in studying the genetics of behavior, an important question is whether the combined items or symptoms are indeed genetically similar. If not, combining them will severely dilute the genetic signal, and thus compromise our understanding of underlying genetic mechanisms. In a paper published in Nature Communications (March 2 2018), PhD student Mats Nagel from the Department of Complex Trait Genetics is the first to examine the genetic similarity of individual symptoms of psychological traits in a large-scale study.
The study, led by Sophie van der Sluis from the Department of Complex Trait Genetics of the VU/VUMC, focused on investigating the genetic similarity of 12 symptoms generally used as indicators of the personality trait neuroticism, using a sample of 375,913 individuals from the UKBiobank cohort. Neuroticism is a stable personality trait that correlates with psychiatric traits like anxiety, substance abuse, and major depressive disorder. Through extensive analyses and function annotation, Nagel and colleagues showed that the 12 items are genetically quite different, and that important biological understanding can be gained by analyzing the items individually rather than focusing only on their sum. Interestingly, 2 subclusters of genetically very similar items could be identified, reflecting ‘depressed affect’ and ‘worry’. The genetic distinctness of these subclusters was supported by analyses quantifying their genetic relatedness to other traits like BMI, educational attainment, and ADHD.
Furthermore, Nagel et al identified 255 genome-wide significant independent genomic regions, of which 138 were symptom-specific. They found a set of genes that were associated with all symptoms, but also genes that were more symptom-specific. These results indicate that specific symptoms of the same trait may be influenced by different pathways, which will go undetected when investigating a composite score. The current study identifies several of the symptom specific genetic routes emphasizing the genetic heterogeneity of neuroticism.
As a first of its kind, this symptom-level study highlights the purpose and relevance of item- or symptom-level analyses as a means to further our understanding of the genetics of complex behavior.
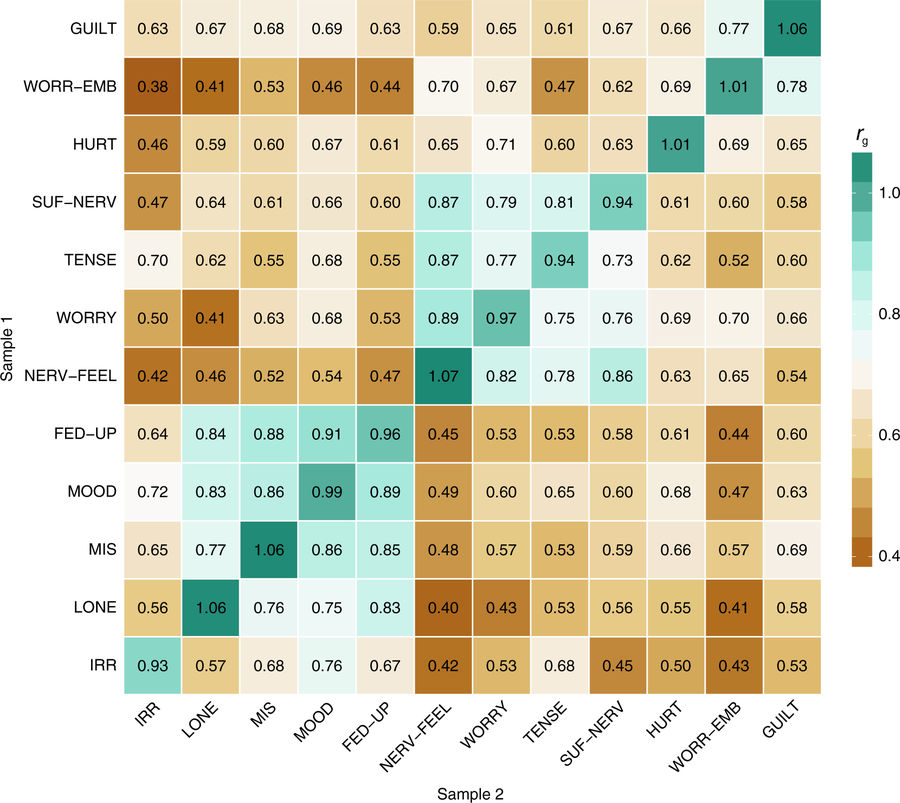
Fig. Genetic correlations between the 12 neuroticism items
Genetic correlations, computed with bivariate LD score regression, between the 12 Neuroticism items in two samples (Sample 1: upper triangle, Sample 2: bottom triangle). On the diagonal are the genetic correlations of the same items measured in both samples. In green, the 2 subclusters consisting of 4 items each. Cluster 1 was denoted Depressed Affect, cluster 2 was denoted Worry.
The study was funded by a VIDI grant from the Netherlands Organization for Scientific Research awarded to Sophie van der Sluis. It is published in Nature Communications, March 2, 2018