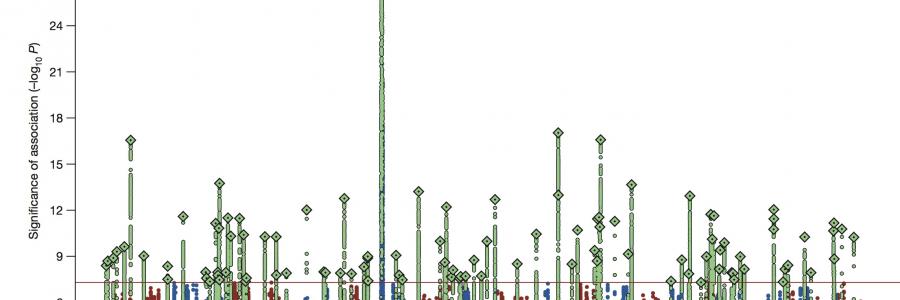
In a study published in Nature, researchers from CTGlab have helped identify over 100 genetic variants for schizophrenia in what is the largest genomic study published on any psychiatric disorder to date.
Schizophrenia, a debilitating psychiatric disorder that affects approximately 1 out of every 100 people worldwide, is characterized by hallucinations, paranoia, and a breakdown of thought processes, and often emerges in the teens and early 20s. Its lifetime impact on individuals and society is high, both in terms of years of healthy life lost to disability and in terms of financial cost, with studies estimating the cost of schizophrenia at over $60 billion annually in the U.S. alone.
Despite the pressing need for treatment, medications currently on the market treat only one of the symptoms of the disorder (psychosis), and do not address the debilitating cognitive symptoms of schizophrenia. In part, treatment options are limited because the biological mechanisms underlying schizophrenia have not been understood. The sole drug target for existing treatments was found serendipitously, and no medications with fundamentally new mechanisms of action have been developed since the 1950s.
In the genomics era, research has focused on the genetic underpinnings of schizophrenia because of the disorder’s high heritability. Previous studies have revealed the complexity of the disease (with evidence suggesting that it is caused by the combined effects of many genes), and roughly two dozen genomic regions have been found to be associated with the disorder. The new study confirms those earlier findings, and expands our understanding of the genetic basis of schizophrenia and its underlying biology.
In the genome-wide association study (GWAS) published in Nature, the authors looked at over 80,000 genetic samples from schizophrenia patients and healthy volunteers and found 108 specific locations in the human genome associated with risk for schizophrenia. Eighty-three of those loci had not previously been linked to the disorder.
“The progress in this field that we have made in just a few years is unprecedented. Just five years ago, there were less than a handful of reliable, replicable genetic variants for schizophrenia, and now, in an enormous collaborative effort, we are able to identify hundreds of them”, said co-author Danielle Posthuma, a scientist at the VUMC and VU Center for Neurogenomics and Cognitive Research, who has been involved in the PGC study from its inception. “Apart from identifying single genetic variants, we are now able to connect the multiple variants and discover genetic pathways important to schizophrenia, which allows us for the first time to begin to understand the disorder at the cellular level.”
The study implicates genes expressed in brain tissue, particularly those related to neuronal and synaptic function. These include genes that are active in pathways controlling synaptic plasticity – a function essential to learning and memory – and pathways governing postsynaptic activity, such as voltage-gated calcium channels, which are involved in signaling between cells in the brain.
Additionally, the researchers found a smaller number of genes associated with schizophrenia that are active in the immune system, a discovery that offers some support for a previously hypothesized link between schizophrenia and immunological processes. The study also found an association between the disorder and the region of the genome that holds DRD2 – the gene that produces the dopamine receptor targeted by all approved medications for schizophrenia – suggesting that other loci uncovered in the study may point to additional therapeutic targets.
The study is the result of several years of work by the Schizophrenia Working Group of the Psychiatric Genomics Consortium (PGC, http://pgc.unc.edu), an international, multi-institutional collaboration founded in 2007 to conduct broad-scale analyses of genetic data for psychiatric disease. A total of 55 datasets from more than 40 different contributors were needed to conduct the analysis. The Dutch Genetic Cluster Computer, funded by the VU and hosted by SurfSARA provided the necessary storage and computing power for the study.
“The level of data used to conduct the study is massive and required access to high-performance computing. The Genetic Cluster Computer played an essential role in facilitating the necessary scale of analysis for the study. The study would have taken multiple additional years to complete without the Dutch Cluster”, said Danielle Posthuma, who coordinates access and analyses on the cluster for the PGC.
The 80,000 samples used in this study represent all of the genotyped datasets for schizophrenia that the consortium has amassed to date. The PGC is currently genotyping new samples to further study schizophrenia and additional psychiatric diseases, including autism and bipolar disorder.
Core funding for the Psychiatric Genomics Consortium comes from the U.S. National Institute of Mental Health (NIMH), along with numerous grants from governmental and charitable organizations, as well as philanthropic donations. The Genetic Cluster Computer is funded by the VU University, with a supplement from the Dutch Brain Foundation, the Netherlands Organization of Scientific Research, as well as donations from several international research groups.