PhD student Kyoko Watanabe (CNCR-CTG) curated 4155 GWAS results and analyzed558 traits to explore fundamental genetic questions. The study is published in Nature Genetics.
A decade of genome-wide association studies (GWAS) has provided a wealth of genetic associations for hundreds of human traits. In this new study, the team, led by Danielle Posthuma of the VU University Amsterdam and Amsterdam UMC, systematically analyzed the results of virtually all published GWAS results. All results are available through a novel central database gwasATLAS.
“Our study provides insight into fundamental questions on the extent of pleiotropy across the genome, the nature of risk variants and the genetic architecture of complex traits.” says Kyoko Watanabe, a PhD student in the group of Posthuma and the first author on the study.
Many genes influence multiple traits
‘Pleiotropy’ refers to the observation that one gene can influence multiple traits. “Many traits are known to be influenced by hundreds of genes, and given that we have a finite number of genes in our genome, it has been hypothesized that pleiotropy is ubiquitous in our genome. In the current study we quantified this, by analyzing the results of well-powered GWAS studies for more than 500 different traits. “ explains Posthuma.
More than half the genome was found to be associated with at least one trait, and
66% of 17.5k tested genes were associated with at least one trait. Of these associated genes, 81% were associated with multiple traits. A specific region on chromosome 3 harbored several highly pleiotropic genes, such as TRAIP, CAMKV, MST1R, MON1A and RBM6.
“These results show that pleiotropy is the rule rather than the exception, and that one gene can be involved in hundreds of different traits. Thus, variations in one gene may for example be associated with a slight increase in risk of being bald, a tiny decrease in the risk of being neurotic, and may add 0.02 cm to a person’s height”, says Watanabe.
The finding that the majority of genes are influencing multiple traits suggests that strategies such as gene-editing for complex traits based on GWAS results can be risky. “One problem is that GWAS results do not provide causal variants, but provide a range of variants that might include the causal variant. A second problem is that to influence one trait via gene-editing, one would need to edit hundreds of variants in hundreds of genes, and since these genes may not only influence the targeted trait, unintended effects on hundreds of other traits may occur.” says Posthuma.
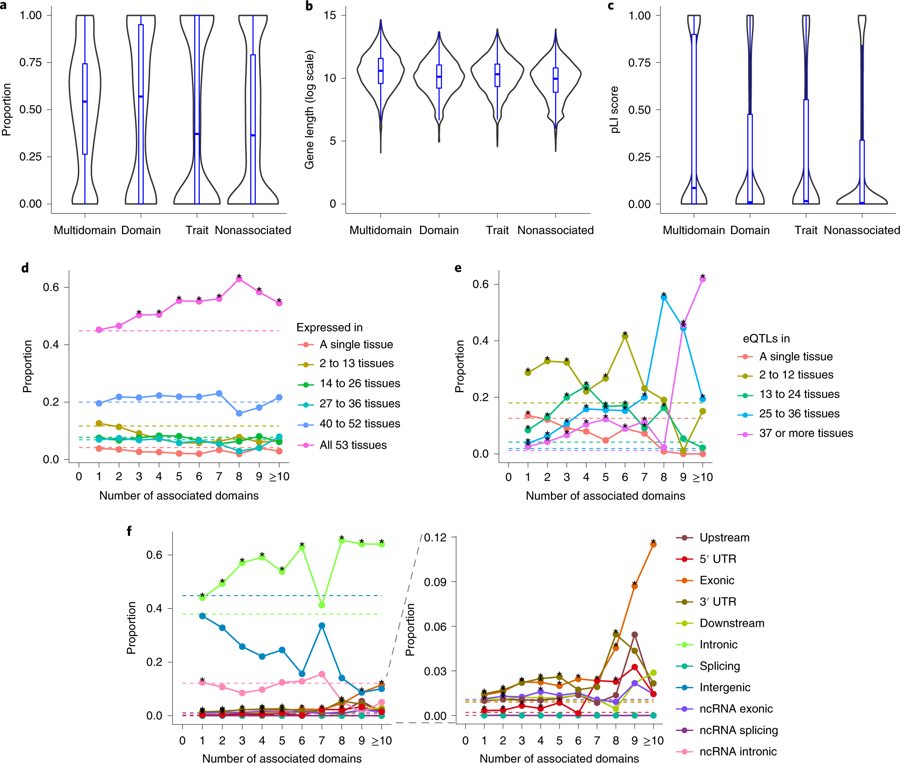
a, Distribution of gene density of loci with different association types. b, Distribution of gene length in log scale with different association types. c, Distribution of pLI scores of genes with different association types. a–c, Multidomain: associated with traits from >1 domain, domain: associated with >1 trait from a single domain, trait: associated with a single trait, nonassociated: not associated with any of 558 traits. d, Tissue specificity of genes at different levels of pleiotropy. Each data point represents a proportion of genes expressed in a given number of tissues for a specific number of associated domains. e, Proportion of SNPs with different functional consequences at different levels of pleiotropy. Each data point represents the proportion of SNPs with a given functional consequence for a specific number of associated domains. f, Tissue specificity of SNPs based on active eQTLs at different levels of pleiotropy. Each data point represents the proportion of SNPs being eQTLs in a given number of tissues for a specific number of associated domains. d–f, Dashed lines refer to the baseline proportions (relative to all 17,444 genes (d) or all 1,740,179 SNPs (e,f)) and stars denote significant enrichment relative to the baseline (one-sided Fisher’s exact test). (From Watanabe et al., 2019)
Trait-specific genes are most interesting for drug development
The results of the study showed that genes associated with multiple traits have a significantly higher probability of being intolerant to mutations that would shut them down completely, and that these genes are expressed in more tissue types compared to other genes. “This suggests that genes associated with multiple traits tend to have more general biological functions, indicating that the bulk of genes is involved in a general susceptibility to trait variation and variation in risk to disease.” says Watanabe.
The study also found several genes which were specific to one disease or trait. “These genes are highly informative for that particular trait, and therefore the most interesting targets for functional follow-up studies and drug developments”, says Posthuma
Some traits are extremely polygenic
The study also reports on the expected sample sizes needed to detect all involved causal genetic variants each of the investigated traits. Some traits, like major depressive disorder and insomnia, were found to be extremely polygenic. “For these traits we need tens of millions of individuals to detect 90% of all causal variants”, adds Watanabe.
The study has been carried out in collaboration with the teams of Ben Neale from the Broad Institute, Cambridge, US and Ole Andreassen from the University of Oslo, and was made possible thanks to the fantastic resources of UKBiobank (https://www.ukbiobank.ac.uk) and computing resources provided by Surfsara (https://userinfo.surfsara.nl/systems/lisa) and funding provided by NWO-VICI.
The study is published as:
Kyoko Watanabe, Sven Stringer, Oleksandr Frei, Maša Umićević Mirkov, Christiaan de Leeuw, Tinca J. C. Polderman, Sophie van der Sluis, Ole A. Andreassen, Benjamin M. Neale & Danielle Posthuma. A global overview of pleiotropy and genetic architecture in complex traits. Nature Genetics, DOI:10.1038/s41588-019-0481-0
Online visualization and exploration of thousands of GWAS results:
gwasATLAS.