A collaboration between Marijn Schipper and Danielle Posthuma from CNCR-CTG and Camiel Mannens and Sten Linnarsson from the Karolinska Institute Sweden has led to the publication of the first map of chromatin accessibility and paired gene expression in the entire developing early embryonic human brain. The study is published May 1, 2024 in Nature.
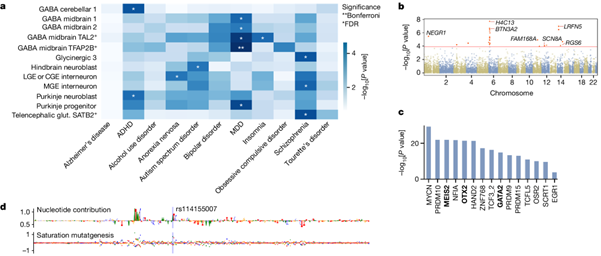
The high-resolution multi-omic atlas of chromatin accessibility and gene expression during the first-trimester of the developing human brain includes more than 100,000 cell-type- and region-specific developmental accessible chromatin regions, as well as inferred candidate cis-regulatory elements and their predicted regulatory effects. The resource allows for example linking transcription factors to putative enhancers, and enhancers to their target genes. In addition, this resource enables the interpretation of genetic associations with disease.
Marijn Schipper, PhD student at CNCR-CTG, investigated whether the parts of DNA that contain open chromatin in specific types of cells during early embryonic development overlap with genetic variants that have been identified previously in genomewide association studies for psychiatric disorders. We found that the patterns of open chromatin in the DNA in specific types of cells during development coincide with stretches of DNA that are also linked to ADHD, anorexia, autism spectrum disorders, depression, insomnia and schizophrenia. The strongest association was found for depression: genetic variants that have previously been linked to depression occur particularly frequently in stretches of DNA that are mainly active during early embryonic development in the GABAergic neurons in the midbrain. This link with the midbrain is novel, and offers new research directions with a specific focus on the midbrain for understanding the hereditary component of depression.
The paper can be found here: https://www.nature.com/articles/s41586-024-07234-1