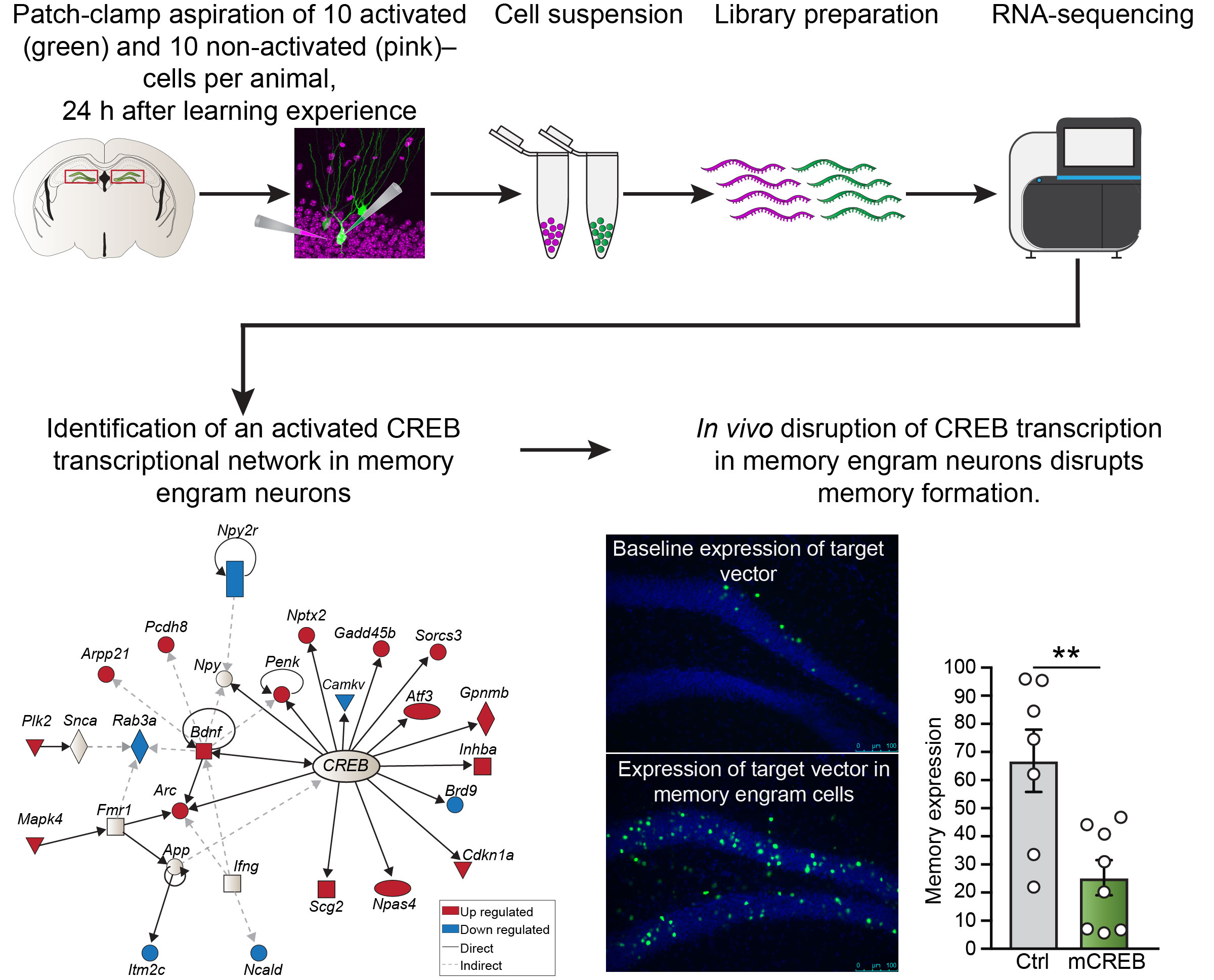
A team of researchers from the CNCR (led by Priyanka Rao-Ruiz and Michel van den Oever, MCN lab) and the Erasmus MC (led by Steven Kushner, Dept. of Psychiatry) used a sensitive and unbiased RNA sequencing approach to identify gene expression signatures critical to the storage of long-term memories by sparse ensembles of hippocampal neurons. This collaborative effort was published in Nature Communications.
“A learning experience activates small populations of neurons in the brain’s hippocampus and these activated neurons collectively form a memory ensemble or engram. This newly acquired memory must then be stabilized for long-term storage by a time-dependent process of consolidation that requires persistent molecular adaptations in engram neurons. However the identity of these molecular signatures of memory have remained largely unknown thus far due to the technical difficulty in probing these sparsely distributed neurons at a molecular level.” says Priyanka Rao-Ruiz, first author of the study.
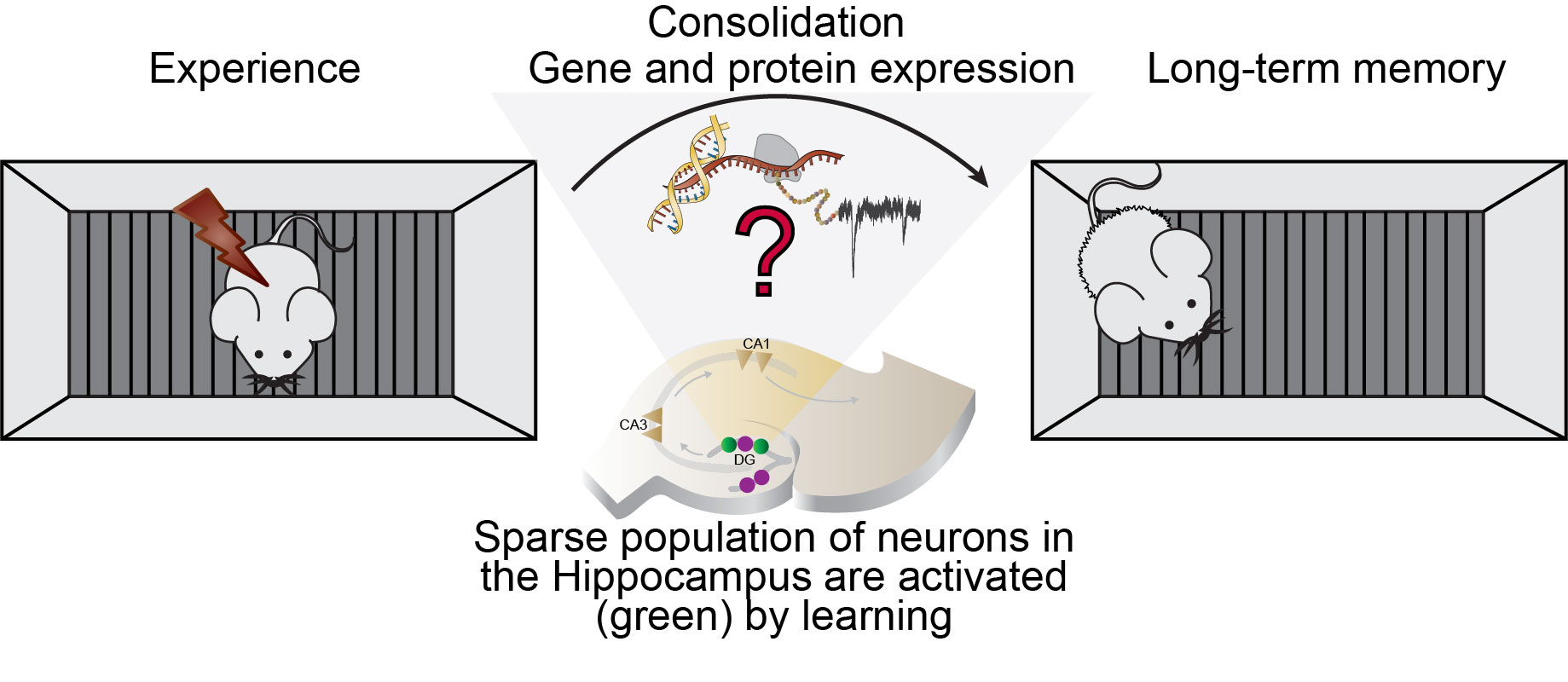
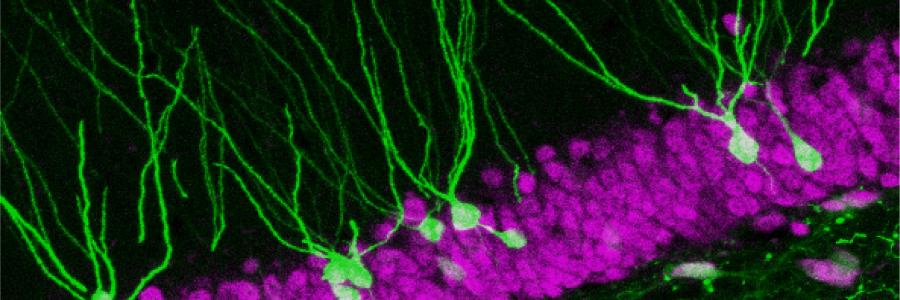
To tackle this issue, researches used activity-dependent transgenic reporter mice in combination with in vivo miniature microendoscopy to express a fluorescent tag in neurons activated by a specific learning experience (fear conditioning) and visualize them over time. A novel and sensitive method of fluorescence-guided cell aspiration was then used to harvest and selectively isolate the content of 10 activated hippocampal neurons for sensitive RNA-sequencing. A comparison between the molecular profile of memory engram neurons and their non-activated neighbors 24 h after a learning experience revealed enduring molecular modifications specific to memory consolidation. In particular, a network of genes whose expression is modulated by a multifaceted regulator of gene expression (i.e. CREB), featured prominently. Finally, the functional relevance of these changes in gene expression was confirmed by engram-specific disruption of the identified CREB gene network using a novel viral vector designed by the team of Michel van den Oever. This targeted in vivo manipulation resulted in a profound deficit in memory storage and its subsequent expression.
Together, this study has identified critical molecular mechanisms necessary for experience-dependent plasticity that drives the consolidation of long-term memories by sparse neuronal ensembles. This study was funded in part by a ZonMW TOP grant. Download the original article here.
Read more about the Memory Circuits team and Molecular Engrams team