A new study by PhD student Martina Rosato (MCN) shows that combined cellular and molecular profiling is able to reveal novel biological insight into the polygenic nature of schizophrenia. The study was published last month in Molecular Psychiatry.
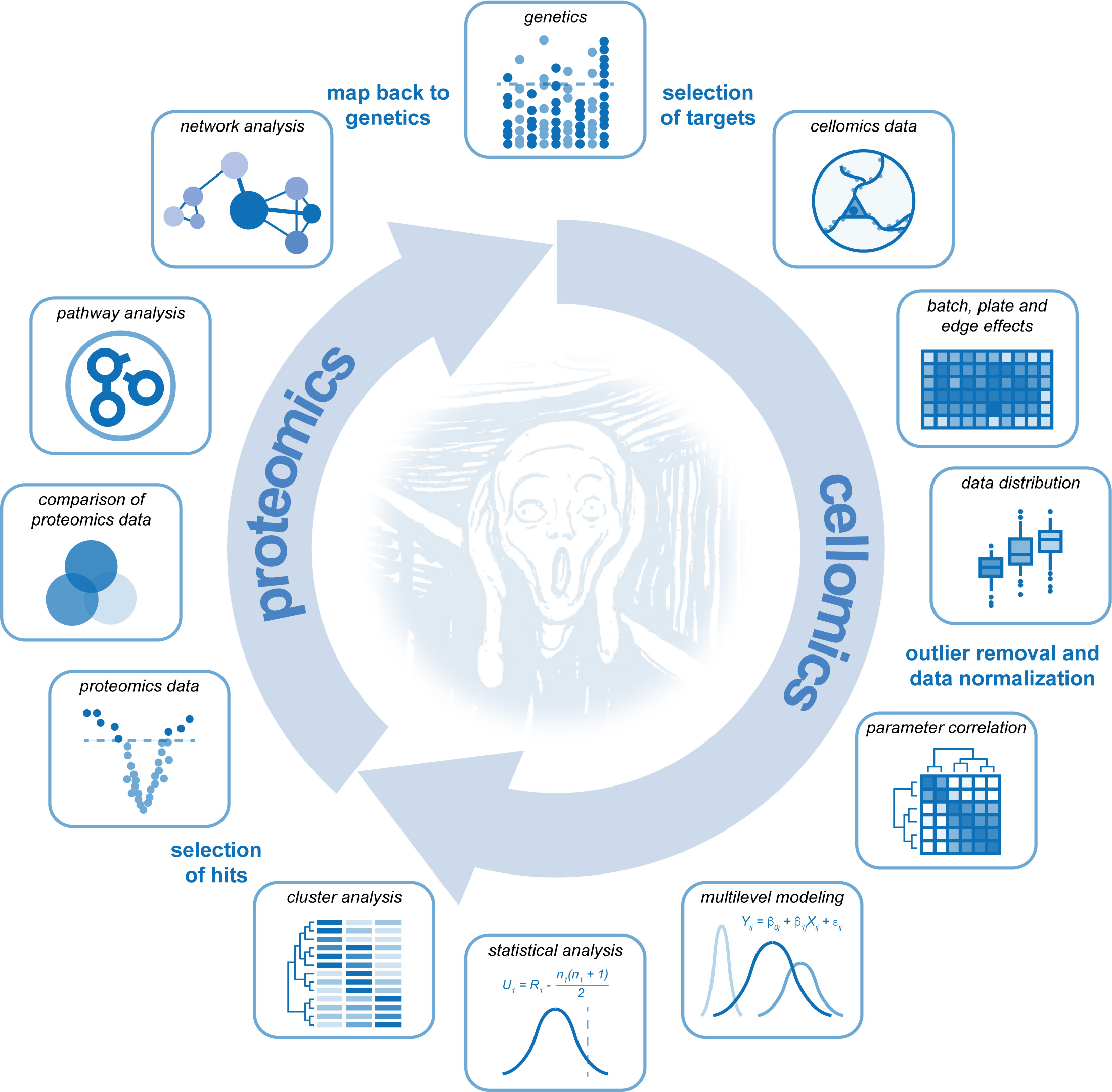
Schizophrenia is highly polygenic in nature with more then 145 identified risk loci. It is generally assumed that multiple risk-increasing mutations are required to get schizophrenia, but how risk genes act together on brain systems remains elusive. Geneticists mainly rely on computational tools in order to predict polygenic risk of particular gene combinations. In this paper, Martina Rosato took advantage of high-throughput cellular screening technologies in order to find experimental evidence for the functional convergence and combined action of multiple schizophrenia risk genes. She systematically knocked down the expression of many predicted risk genes one-by-one in mouse embryonic primary neurons and classified them according to the cellular phenotype that they produced. Genes that produced the same phenotype were then hypothesized to act in common neuronal pathways to potentially increase neuronal dysfunction in a synergistic manner. This hypothesis was subsequently tested for three genes, Tcf4, Tbr1 and Top3b, which al reduced synapse numbers and for which a common synaptic function would not have been predicted a priori. Martina performed a molecular (proteomics) analysis of neurons in which the expression of either of these three genes was knocked down, and found that all three alter the expression certain components of the neurotransmitter release machinery. Interestingly, their individual impact on neurotransmitter release was predicted to be small and insignificant, however, for the three genes combined this effect was highly significant. Together, these data support the idea that polygenic risk is the result of multiple small impacts on common neuronal signaling pathways, and that interpreting polygenic risk from a protein regulatory network perspective is able to uncover hidden aspects of disease biology.
This study was part of the SUN project coordinated by UNC geneticist Patrick Sullivan and funded by the Karolinska Institute (Stockholm, Sweden), and performed in collaboration with CNCR geneticist Daniëlle Posthuma (CTG). A full-text pdf of the paper is available here.